Lazy hacker's service analytics
A week ago, I had trouble with the GHTorrent data retrieval process. Specifically, while scripts where performing as expected and the event processing error rate was within reasonable bounds, API requests took forever to complete, in many cases as much as 20 seconds. I know that Github’s API is very snappy, and even though it the response times I get are slower than what Github reports, it is reasonably fast if we take into account the packet trip over (or under) the Atlantic (usually, around 500msec).
My main hypothesis was that Github started employing some kind of tar pitting strategy for accounts using their API extensively. I am one of them: for every Github account that I have collected from fellow researchers, I run two mirroring processes, to make sure that I exploit the full 5000 requests/sec limit. As I maintain extensive (debug-level) logs for each request GHTorrent makes, I decided to investigate whether that was the case.
Preparing the data
I had to process > 10 GB of data on a 16-core server and extract information
about URLs and timings. This was a non-brainer: Unix all the way! I slapped
together the following (totally inefficient) script, which uses convenient
functions in 3 programming languages :-) The crucial thing here is the use of
the parallel command. This allows the doit() function to be applied in
parallel on 10 input files, thereby stressing the fast machine sufficiently
enough.
The script outputs a CSV file with 3 fields: timestamp (as seconds since the epoch), IP address used for the request, time the request had taken.
processfile() {
grep APIClient $1|
grep -v WARN |
perl -lape 's/\[([T0-9-:.]*).*\] DEBUG.*\[([0-9.]*)\].*Total: ([0-9]*) ms/$1 $2 $3/'|
cut -f2,3,4 -d' '|
ruby -ne 'BEGIN{require "time"}; t,i,d=$_.split(/ /); print Time.parse(t). to_i," ", i, " ", d;'|
grep -v "#"
}
export -f processfile
find mirror -type f|grep log.txt| parallel -j10 processfile {}Data processing with R
I have a love-hate relationship with R. I do admire the fact that it allows developers to acquire data from multiple sources, manipulate it relatively easily and plot it beautifully. Also, whenever basic R has shortcomings, the community usually steps is with awesome libraries (be it sqldf, plyr, ggplot2 etc). R the language, however, leaves much to be desired. Nevertheless, as I had written lots of R code lately, it somehow felt like an obvious choice for summarizing the data. Here is the script I came up with.
Importing tabular data in R is trivial: a call to read.csv will do the job
without any trouble, loading a comma or tab separated file in an in-memory
representation called a data frame. I usually also pass to it column type
parameters to make sure that integers are indeed represented as integers and
factors are recognized as such, and also to make sure that there are no errors
in the data file. Moreover, to have a flexible representation of time, I usually
convert epoch timestamps to the POSIXct data type.
After initial importing of the file, basic statistics (quantiles and means) can be acquired using the summary function.
Typical processing of time series data includes aggregation per a configurable time unit (e.g. hour, day etc). In R, this can be achieved using a two step process: i) binning (assigning labels to data based on a criterion) ii) bin-based aggregation. Fortunately, both steps only consist of a line each! For example, if we want to aggregate the mean time of each API request per 30 minutes it suffices to do the following:
data$timebin <- cut.POSIXt(data$ts, breaks = "30 mins")
count.interval <- aggregate(ms ~ timebin, data = data, mean)cut.POSIXt is an overload of the general cut binning function that works on
data of type POSIXt. As it knows that it works on time data, the binning is
time aware, so we can specify arbitrary time-based bins (e.g. ‘12 minutes’ or ‘3
months’). The aggregate function will then summarize our data given a formula:
in our case, it will apply the aggregation function mean on groups of ms,
where the grouping factor is the assigned time bin. In SQL, the equivalent
expression would be: SELECT timebin,mean(ms) FROM... GROUP BY timebin. We can
pass multiple grouping factors and arbitrary aggregation functions, which makes
aggregate’s functionality a superset of SQL, for single tables. For more
complex aggregations (e.g. self joins), we can use sqldf to convert our data
frame in an SQLite table, run an SQL query and get back a new data frame with
the results in one go.
The final step would be to plot the aggregated data. For that, I use ggplot2,
which, in my opinion, is the best plotting tool bar none. Using ggplot2 is
straightforward, after one understands the theory behind
it. In the
following example, we specify an aesthetic (roughly, the data to be plotted)
which we then use to feed a line plot with a date x-axis. ggplot2 will take
care of scales, colors etc.
ggplot(mean.interval) +
aes(x = timebin, y = ms) +
geom_line() +
scale_x_datetime() +
xlab('time') +
ylab('Mean API resp in ms') +
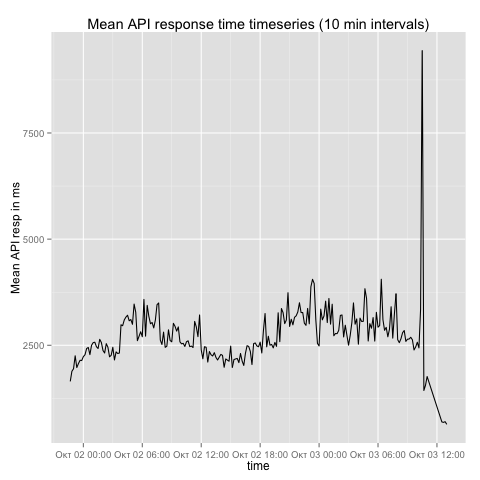
ggtitle('Mean API response time timeseries (10 min intervals)')The plot that resulted from the R script on the data I processed can be seen below:
Results
After I had the evidence at hand, I had a brief discussion with our network administrators. They had recently updated the university-wide firewall policies for higher throughput. Unfortunately, this turned out to be at the expense of latency. As we can see at the end of the plot above, after the changes where reverted the mirroring process started flying again, with 500 msec average latency. So Github was innocent and my working hypothesis wrong.
The net result is that with 40 lines of totally unoptimized code, I can go through several gigabytes worth of logs and plot service quality timeseries plots in a little over a minute. Since I had the initial implementation running reliably, I created a few more plots and added a cron job and a web page to display them online. You can also see them here.
The moral of the story is that we don’t always need to setup complicated systems to do service monitoring. A few Unix commands to extract data from logs and a tool to summarize and plot them might be enough to get us going.